- Marine Glycobiology
- Research
Research

Marine carbon cycle: a glycobiology perspective
Research in our laboratory focuses on how microbes, pathways and enzymes control the turnover of algal carbohydrates. The oceans store significant amounts of carbon dioxide locked within polysaccharides, which belong to the most abundant, diverse and complicated molecules within the marine carbon cycle. They are the major output of photosynthesis, provide energy and carbon for marine food webs and act as a potential carbon sink in the global cycle. Microbes hold the key to unlock the carbon energy stored in polysaccharides. They have the enzymes to rapidly deconstruct and consume polysaccharides. We have shown that the enzymes and pathways for the degradation of polysaccharides transfer easily between microbes enabling them to explore new food sources through a process called horizontal gene transfer. However, the high diversity of polysaccharides, which enables microbes to specialize, also poses a problem for carbon turnover. We propose that the high structural complexity of polysaccharides renders them intrinsically recalcitrant against microbial degradation and makes them a potential carbon sink in the marine carbon cycle. Polysaccharides have highly complicated three dimensional structures that require microbial pathways with many unique enzymes for their complete degradation. To explore these ideas we use classical omics technologies, biochemistry, structural biology and apply established technologies from biomedicine in the ocean. In essence, we learn from microbes to study the marine carbon cycle.
Microbial utilization of carbohydrates
The identity and molecular degradation pathways of marine carbohydrates remain obscure. We use marine bacterial isolates that degrade key carbohydrate polymers as model organisms to resolve the mechanistic details. The molecular pathways involved in polysaccharide utilization are analyzed on three levels: 1) Microbial physiology experiments with purified marine polysaccharides reveal the substrate level details of a strain’s niche. 2) Proteome analysis is used to elucidate the machinery involved in degrading the high molecular weight compounds. 3) Individual enzymes and carbohydrate binding proteins involved in the pathways are recombinantly expressed, purified and subjected to biochemical characterization and structural characterization using X-ray crystallography. Overall, the combination of these techniques provides a robust platform for a molecular level understanding of the marine carbohydrate cycle.

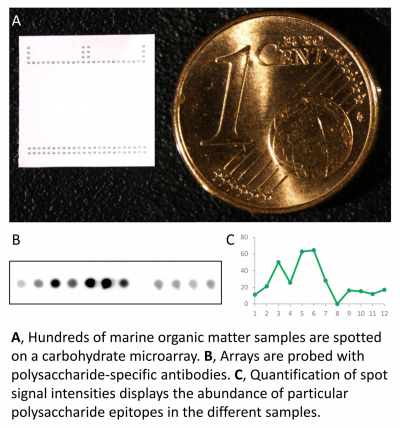
Environmental glycomics
Microalgae provide the nutritional foundation of marine food webs. They create about 45 gigatonnes of organic carbon per year, a major fraction is in the form of polysaccharides. However, we know very little about what types of polysaccharides are produced and how fast they are catabolized in the environment. Polysaccharide structures are challenging to analyse with current methods, owing to their complexity. They contain numerous monosaccharides with diverse linkages and are often modified with a range of chemical groups. Individual marine polysaccharides are often in low abundance and are hidden within a complex mixture. As such, current analytical methods cannot resolve individual polysacharides because they require large amounts of purified substrate. We experiment with carbohydrate microarrays, a technique established in the biomedical and land plant research fields. This technology combines the high specificity of antibodies with the throughput of microarrays to identify and follow the turnover of polysaccharide structures in the ocean.
Molecular structure and reactivity
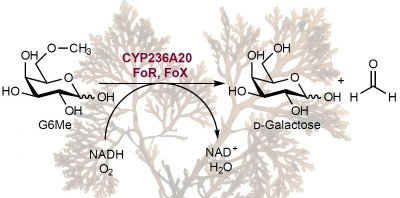
Marine polysaccharides are thought to be fast-food for microbes. However, polysaccharides can also accumulate in seawater and sediments. We investigate factors that influence the reactivity of polysaccharides and ask whether they can be a sink in the global carbon cycle. Our aim is to link polysaccharide structure to its turnover rate by microbes. We use defined polysaccharides and cultivated isolates to identify limitations for fast polysaccharide degradation. The ability of microbes to digest those substrates is probed with genomics, proteomics, physiological experiments and enzyme assays in the laboratory and in the environment.
Tools for marine glycobiology
Marine polysaccharides have high structural diversity and are notoriously difficult to analyze or quantify in natural samples. This challenges conventional analytic techniques and requires additional molecular tools for their analysis. We rely on evolutionary principles to solve this problem. Microbes evolved to overcome the diversity of polysaccharides with specific enzymes and carbohydrate binding proteins. We exploit these specific proteins from bacterial genomes or metagenomes and use them to identify and quantify polysaccharides (the principle is described in our recent paper). Aside from this primary application in environmental science, these enzymes also have biotechnological potential because they convert abundant polysaccharides into fermentable sugars for biofuels and other applications. We also share the proteins with the scientific community through Addgene.
© S. Becker, MARUM
Microbial adaptation and pathway evolution
Marine polysaccharides are complex, diverse and difficult to utilize. Microbes need complex utilization pathways to degrade, transport and metabolize them. Polysaccharide utilization pathways are frequently exchanged between microbes by horizontal gene transfer. We recently showed that those pathways are exchanged even between organisms from different ecosystems. This creates an opportunity to study adaptation and evolution of pathways in microbes. We combine bioinformatics and biochemistry to elucidate how the pathways adapt to their new environment. We develop new computational tools to predict pathways in genomes and metagenomes. This may reveal how carbohydrate resource partitioning contribute to microbial evolution and ecology.