Die Inhalte dieser Seite sind leider nicht auf Deutsch verfügbar.
Seitenpfad:
- Organische Geochemie
- Techniken
- Hinrichs Lab - Lipid Biomarkers
Hinrichs Lab - Lipid Biomarkers
Introduction
Lipid biomarkers are specific organic molecules that can be traced back to their natural source organism in the environment. Biomarkers are detected in natural samples as structurally intact or rearranged biomolecules thus characterizing biomass of modern and ancient (micro)organisms. The Hinrichs Lab analyzes all kinds of organic molecules found in environmental samples ranging from tradtional (GC-amenable) biomarkers to tetraether lipids (core lipids) to intact polar membrane lipids (IPLs).

Traditional biomarkers
The group of traditional biomarkers in organic geochemistry is very diverse and includes compounds from a wide range of polarities and molecular structures stretching from hydrocarbons to fatty acids and from acyclic n-alkanes and isoprenoids to cyclic steroidal lipids or hopanoids.
Analysis of biomarkers is performed via gas chromatographic separation based on differences in volatility, i.e. boiling points, and their interaction with the stationary phase of the capillary column and the mobile gas phase, classically being apolar columns and helium, respectively. Whereas apolar compounds such as hydrocarbons are analyzed directly, compounds with polar functional groups have to be derivatized before analysis on these columns due to otherwise bad peak shapes and low detection. Biomarkers are detected by flame ionization (FID) and/or mass spectrometry (MS). The former is dominantly used for quantification and the latter for structural identification.
We use traditional biomarkers for the investigation of recent and past activities of methane oxidation (Niemann and Elvert, 2008; Wegener et al., 2008; Haas et al., 2010; Cook et al., 2011), the characteristics of marine productivity across past mass extinction events (Sepulveda et al., 2009b), the evaluation of temperature proxies (Leider et al., 2010), and for the study of sources, transport and partioning of organic matter at continental margins (Schmidt et al., 2010). In some cases new biomarker compounds are identified and used as diagnostic markers for organic matter sources and/or biogeochemical processes.
Analysis of biomarkers is performed via gas chromatographic separation based on differences in volatility, i.e. boiling points, and their interaction with the stationary phase of the capillary column and the mobile gas phase, classically being apolar columns and helium, respectively. Whereas apolar compounds such as hydrocarbons are analyzed directly, compounds with polar functional groups have to be derivatized before analysis on these columns due to otherwise bad peak shapes and low detection. Biomarkers are detected by flame ionization (FID) and/or mass spectrometry (MS). The former is dominantly used for quantification and the latter for structural identification.
We use traditional biomarkers for the investigation of recent and past activities of methane oxidation (Niemann and Elvert, 2008; Wegener et al., 2008; Haas et al., 2010; Cook et al., 2011), the characteristics of marine productivity across past mass extinction events (Sepulveda et al., 2009b), the evaluation of temperature proxies (Leider et al., 2010), and for the study of sources, transport and partioning of organic matter at continental margins (Schmidt et al., 2010). In some cases new biomarker compounds are identified and used as diagnostic markers for organic matter sources and/or biogeochemical processes.
We are equipped with multiple GC systems containing various detectors (FID/TCD/PDD) for the quantification of lipid biomarkers but of which some are dedicated for gas analysis. Our GC-MS facility consists of three regular coupled instruments and one online pyrolysis GC-MS system. All of them contain quadrupole mass analyzers with mass ranges up to 1050 enabling identification of most GC-amenable compounds.

Core lipids
Conventional analytical methods, in particular GC, are unsuitable for direct analysis of glyceroldialkylglyceroltetraether (GDGT) lipids because GDGTs are non-volatile high molecular weight compounds. For a long time, the only available method relied on analysis of the GC-amenable products of the ether cleavage reaction (e.g., DeRosa et al., 1977). Hopmans et al. (2000) developed a method that combines normal phase high performance liquid chromatography (HPLC) with positive ion atmospheric pressure chemical ionization mass spectrometry (APCI-MS) for the direct analysis of GDGTs extracted from archaeal cell material and sediments. This method separates GDGT core lipids based on small differences in polarity and sterical configuration on columns based on silica gel modified with amino- or cyano-groups.
The core lipids of IPLs can be studied after hydrolysis of the polar head group (cf. Sprott et al., 1990). First, the total lipid extract (TLE) is separated into apolar and polar fractions with preparative HPLC according to the published method of Biddle et al. (2006) or by silica gel column chromatography (modified from Oba et al., 2006). The apolar fraction containing core lipids can then be directly analyzed with HPLC-APCI-MS, whereas the polar fraction containing IPLs is acid-hydrolyzed to release the core lipids from IPLs.
The core lipids of IPLs can be studied after hydrolysis of the polar head group (cf. Sprott et al., 1990). First, the total lipid extract (TLE) is separated into apolar and polar fractions with preparative HPLC according to the published method of Biddle et al. (2006) or by silica gel column chromatography (modified from Oba et al., 2006). The apolar fraction containing core lipids can then be directly analyzed with HPLC-APCI-MS, whereas the polar fraction containing IPLs is acid-hydrolyzed to release the core lipids from IPLs.
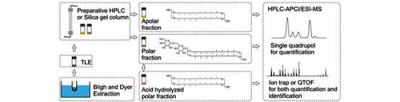
Flow chart showing the procedure of sample preparation and analysis (from Liu, 2011).
For characterization of core lipids, different mass spectrometers are available in the Hinrichs Lab. The selected ion monitoring (SIM) mode of the single quadrupole mass spectrometer is usually used for quantification at high sensitivity, while the ion trap and accurate-mass quadrupole time-of-flight (qTOF) mass spectrometers are ideal for structural identification by providing spectra of parent and fragment (MSn) ions.

Intact polar membrane lipids
After extraction from sediments, pure biomass, or filtered particulate organic matter with a modified Bligh and Dyer method (cf. Sturt et al., 2004), the characterization and identification of IPLs occurs by high-performance liquid chromatography coupled to mass spectrometry. Samples are usually measured in both positive and negative ionization mode to facilitate structural elucidation of compounds. When the mass spectrometer is operating in positive ion mode, the IPL parent ion formed in the ion source typically loses its polar head group upon fragmentation. This results in a characteristic MS2 spectrum from which a head group-diagnostic neutral mass loss can be calculated, e.g., for PE this is 141 Da (cf. Sturt et al., 2004). During fragmentation in negative ion mode the IPL parent ion tends to lose the fatty acids of the hydrophobic side chains which can then be identified according to characteristic fragments in the MS2 spectra, e.g., m/z 255 for the C16:0 fatty acid. A helpful visualization of the data is presented in the form of three dimensional density maps (see Figure). The top panel illustrates an example of a HPLC chromatogram in two dimensions where peaks of individual compounds co-elute and cannot be clearly distinguished. The lower panel shows a three dimensional density map with retention time on the x-axis, the m/z values of the IPL parent ions on the y-axis, and darker colors denoting higher relative intensities in the third dimension. The density map clearly visualizes individual peaks containing different fatty acid side chains and provides a very useful “fingerprint” of the IPL composition of the environmental sample.
The Hinrichs Lab is equipped with several HPLC-MS systems for analysis of intact polar membrane lipids. These systems can either be used for purification of compounds for subsequent characterization (preparative HPLC) or for identification and quantification of individual lipids.

Molecular isotope studies
We analyse the stable carbon isotopic composition of lipid biomarkers such as hydrocarbons, ketones, alcohols and fatty acids separated via GC methods to gain detailed information on source organisms, biogeochemical processes, and the state and perturbations of the carbon cycle (e.g., Niemann and Elvert, 2008; Sepulveda et al., 2009a; Sepulveda et al., 2009b).
We extended this approach towards a combined IPL-CSIA (compound specific isotope analysis) method using preparative-HPLC separating specific IPL groups of which head groups and side-chains are analyzed for stable carbon isotopes after reported cleavage protocols (Lin et al., 2010; Schubotz et al., 2011). This methodology extends the conventional CSIA analysis that is typically performed on bulk IPLs (e.g., Summit et al., 2004; Orcutt et al., 2005; Mills et al., 2006; Wakeham et al., 2007) and has been already applied by Biddle et al. in 2006. Assuming that different types of organisms produce distinct head groups in the same way as side chains this technique has the potential to unravel complex microbial community structures. In concert with the quantification of relative and absolute abundances of IPLs, IPL-specific CSIA can be used to track the carbon flow in various environmental systems.
We extended this approach towards a combined IPL-CSIA (compound specific isotope analysis) method using preparative-HPLC separating specific IPL groups of which head groups and side-chains are analyzed for stable carbon isotopes after reported cleavage protocols (Lin et al., 2010; Schubotz et al., 2011). This methodology extends the conventional CSIA analysis that is typically performed on bulk IPLs (e.g., Summit et al., 2004; Orcutt et al., 2005; Mills et al., 2006; Wakeham et al., 2007) and has been already applied by Biddle et al. in 2006. Assuming that different types of organisms produce distinct head groups in the same way as side chains this technique has the potential to unravel complex microbial community structures. In concert with the quantification of relative and absolute abundances of IPLs, IPL-specific CSIA can be used to track the carbon flow in various environmental systems.
The determination of stable elemental isotopic compositions is a strong tool for the study of various biogeochemical and hydrological processes in the environment. Two of today’s advanced isotope ratio mass spectrometers are available in the Hinrichs Lab and are used for isotope determinations on a wide variety of organic compounds, classes and material.


